Now Reading: Life At The Extremes: Maximally Divergent Microbes With Similar Genomic Signatures Linked To Extreme Environments
-
01
Life At The Extremes: Maximally Divergent Microbes With Similar Genomic Signatures Linked To Extreme Environments
Life At The Extremes: Maximally Divergent Microbes With Similar Genomic Signatures Linked To Extreme Environments

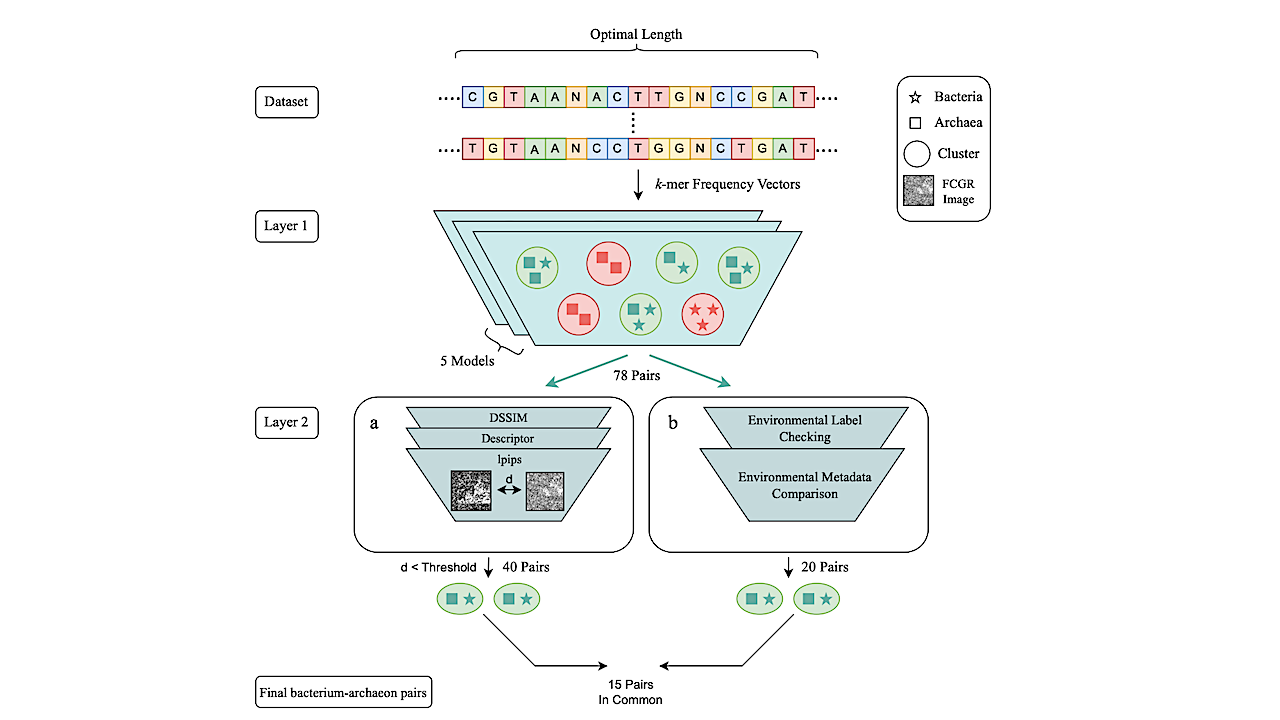
Multi-layered pipeline for identifying bacterium/archaeon pairs with similar genomic signatures. Layer 1: Five selected non-parametric clustering methods identify clusters of organisms with similar genomic signatures. The clusters containing both bacteria and archaea (green) generate a list of 78 candidate bacterium–archaeon pairs, grouped by these algorithms based on their similar genomic signatures. Layer 2-a:: The candidate pairs from Layer 1 undergo pairwise distance calculations between their FCGRs using four different distance metrics. Only 40 pairs, with the majority of distances below empirically determined thresholds, are retained. Layer 2-b: In parallel to FCGR comparison, a biological analysis is conducted on the output pairs from Layer 1. This includes checking environment labels and examining metadata about their living environments to select pairs isolated from similar types of extreme environments, resulting in 20 pairs. The final output is a list of 15 bacterium/archaeon pairs (comprising 16 unique genera and 20 unique species) that passed all filtering layers. These pairs can confidently be proposed as maximally divergent microbes that share similar genomic signatures associated with their living environments. — biorxiv.org
Extreme environments impose strong mutation and selection pressures that drive distinctive, yet understudied, genomic adaptations in extremophiles.
In this study, we identify 15 bacterium–archaeon pairs that exhibit highly similar k-mer–based genomic signatures despite maximal taxonomic divergence, suggesting that shared environmental conditions can produce convergent, genome-wide patterns that transcend evolutionary distance. To uncover these patterns, we developed a computational pipeline to select a composite genome proxy assembled from non-contiguous subsequences of the genome.
Using supervised machine learning on a curated dataset of 693 extremophile microbial genomes, we found that 6-mers and 100 kbp genome proxy lengths provide the best balance between classification accuracy and computational efficiency. Our results provide conclusive evidence of the pervasive nature of k-mer–based patterns across the genome, and uncover the presence of taxonomic and environmental components that persist across all regions of the genome.
The 15 bacterium-archaeon pairs identified by our method as having similar genomic signatures were validated through multiple independent analyses, including 3-mer frequency profile comparisons, phenotypic trait similarity, and geographic co-occurrence data. These complementary validations confirmed that extreme environmental pressures can override traditionally recognized taxonomic components at the whole-genome level.
Together, these findings reveal that adaptation to extreme conditions can carry robust, taxonomic domain-spanning imprints on microbial genomes, offering new insight into the relationship between environmental mutagenesis and selection and genome-wide evolutionary convergence.
Life at the extremes: Maximally divergent microbes with similar genomic signatures linked to extreme environments, biorxiv.org
Astrobiology
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Φsat-2 begins science phase for AI Earth images
03Φsat-2 begins science phase for AI Earth images -
 04Thermodynamic Constraints On The Citric Acid Cycle And Related Reactions In Ocean World Interiors
04Thermodynamic Constraints On The Citric Acid Cycle And Related Reactions In Ocean World Interiors -
 05Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
05Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 06Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
06Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly -
 07Worlds Next Door: A Candidate Giant Planet Imaged in the Habitable Zone of α Cen A. I. Observations, Orbital and Physical Properties, and Exozodi Upper Limits
07Worlds Next Door: A Candidate Giant Planet Imaged in the Habitable Zone of α Cen A. I. Observations, Orbital and Physical Properties, and Exozodi Upper Limits