Now Reading: A Small Polymerase Ribozyme That Can Synthesize Itself And Its Complementary Strand
-
01
A Small Polymerase Ribozyme That Can Synthesize Itself And Its Complementary Strand
A Small Polymerase Ribozyme That Can Synthesize Itself And Its Complementary Strand
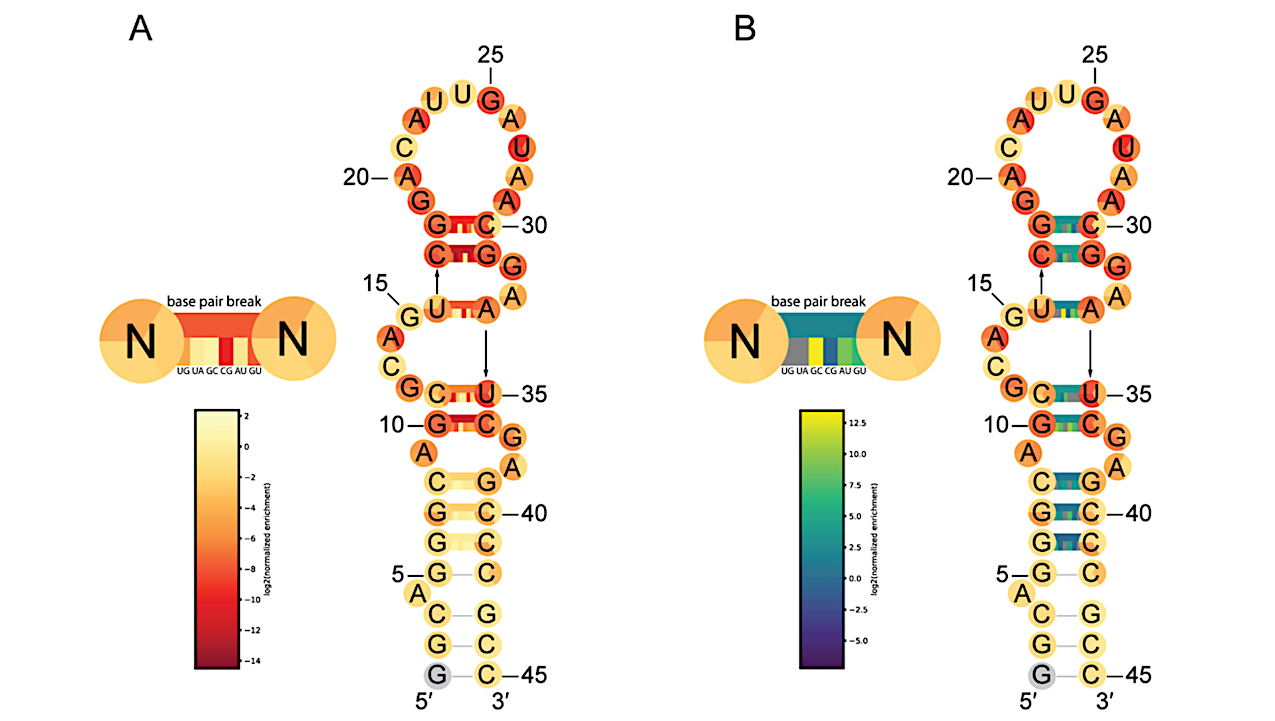

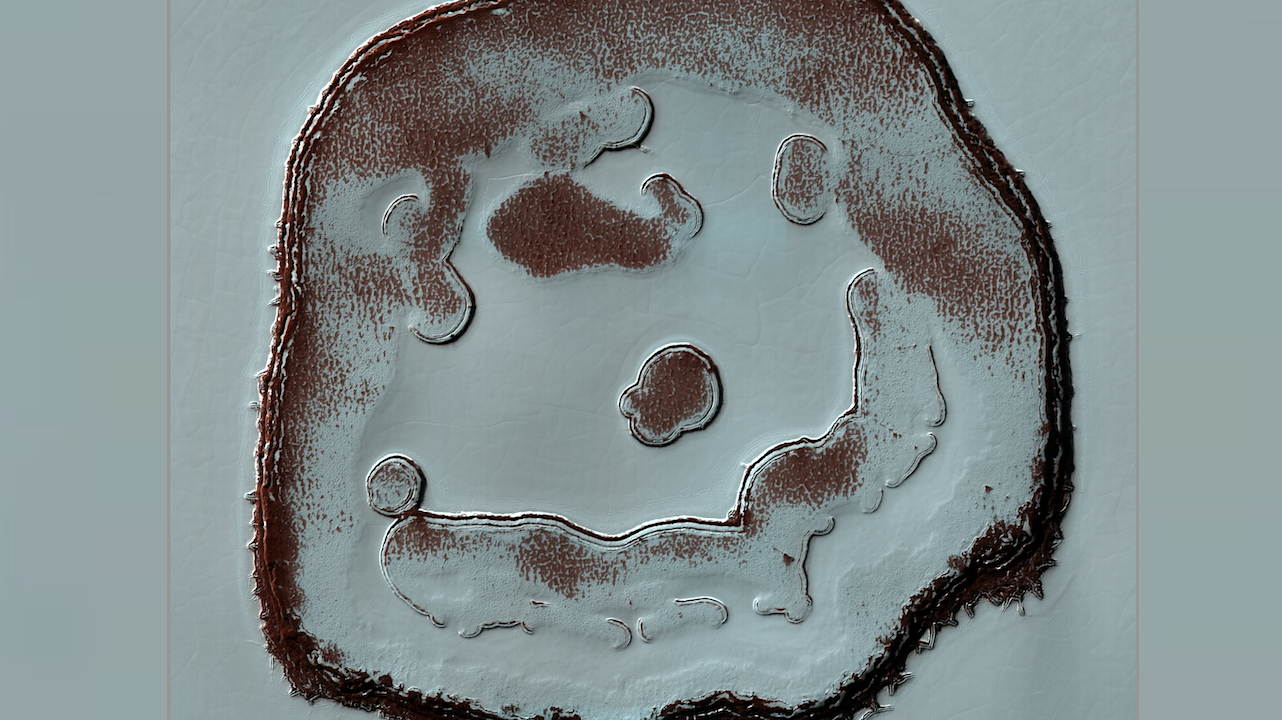
Predicted secondary structure of QT45 ribozyme with nucleotide color corresponding to fitness for each of the three possible single mutations at each position, displayed in the same order as the three representative mutations shown and colored as in Fig. 2E. Base pair annotations are provided, where the upper rectangle connecting two bases shows the average fitness or epistasis of measured base pair-breaking double mutations. For canonical base pairs, the inner small rectangles show (from left to right) the fitness or epistasis of mutations to base pairs UG, UA, GC, CG, AU, GU. For the non-canonical base pair C11-U35, the base pairs shown are UG, UA, GC, UC, CU, CG, AU, GU. (A) Base pair annotations are colored according to fitness. Base pair-retaining mutations generally have higher fitness than base pair-breaking mutations. (B) Base pair annotations are colored according to epistasis. Base pair-retaining mutations generally show more positive epistasis compared to base pair-breaking mutations. — Science
The emergence of a chemical system capable of self-replication and evolution is a critical event in the origin of life.
RNA polymerase ribozymes can replicate RNA, but their large size and structural complexity impede self-replication and preclude their spontaneous emergence. Here we describe QT45: a 45-nucleotide polymerase ribozyme, discovered from random sequence pools, that catalyzes general RNA-templated RNA synthesis using trinucleotide triphosphate (triplet) substrates in mildly alkaline eutectic ice.
QT45 can synthesize both its complementary strand using a random triplet pool at 94.1% per-nucleotide fidelity, and a copy of itself using defined substrates, both with yields of ~0.2% in 72 days. The discovery of polymerase activity in a small RNA motif suggests that polymerase ribozymes are more abundant in RNA sequence space than previously thought.

A small RNA motif with a functionally dense core encodes the triplet polymerase activity of the QT ribozyme. (A) Synthesis of 42 nt CGU repeat sequence by QT51 and its truncation variants. Reaction conditions: 0.25 μM primer F10, 0.25 μM template tP10CGU14, 0.25 μM ribozyme, 5 μM pppCGU triplet, 0.05% Tween 20, 50 mM MgCl2, 50 mM CHES-KOH, pH 9, 5 days at -7 °C frozen. (B) Synthesis of a mixed sequence template by the same ribozymes as in (A). Reaction conditions: 0.25 μM primer F10, 0.25 μM template t6FP10mix, 1.25 μM ribozyme, 5 μM each triplet, 0.05% Tween 20, 50 mM MgCl2, 50 mM CHES-KOH, pH 9, 5 days at -7 °C frozen. In both (A) and (B), ribozymes are not hybridized to template. (C) Predicted secondary structure diagrams of the ribozymes used in (A) and (B) (D) Heatmap of the primer extension activity for all measured single and double mutants of the QT45 ribozyme, with the first constituent point mutation indicated on the x-axis and the second one on the y-axis. Missing datapoints are shown in gray. (E) Predicted secondary structure of QT45 ribozyme with nucleotide color corresponding to its measured average activity on three different templates for each of the three possible mutations, in the same order as the 3 representative mutations provided, indicated in the same color scheme as (D). — biorxiv.org
Astrobiology, Genomics, evolution,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly