Now Reading: Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments
-
01
Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments
Adapting to UV: Integrative Genomic and Structural Analysis in Bacteria from Chilean Extreme Environments
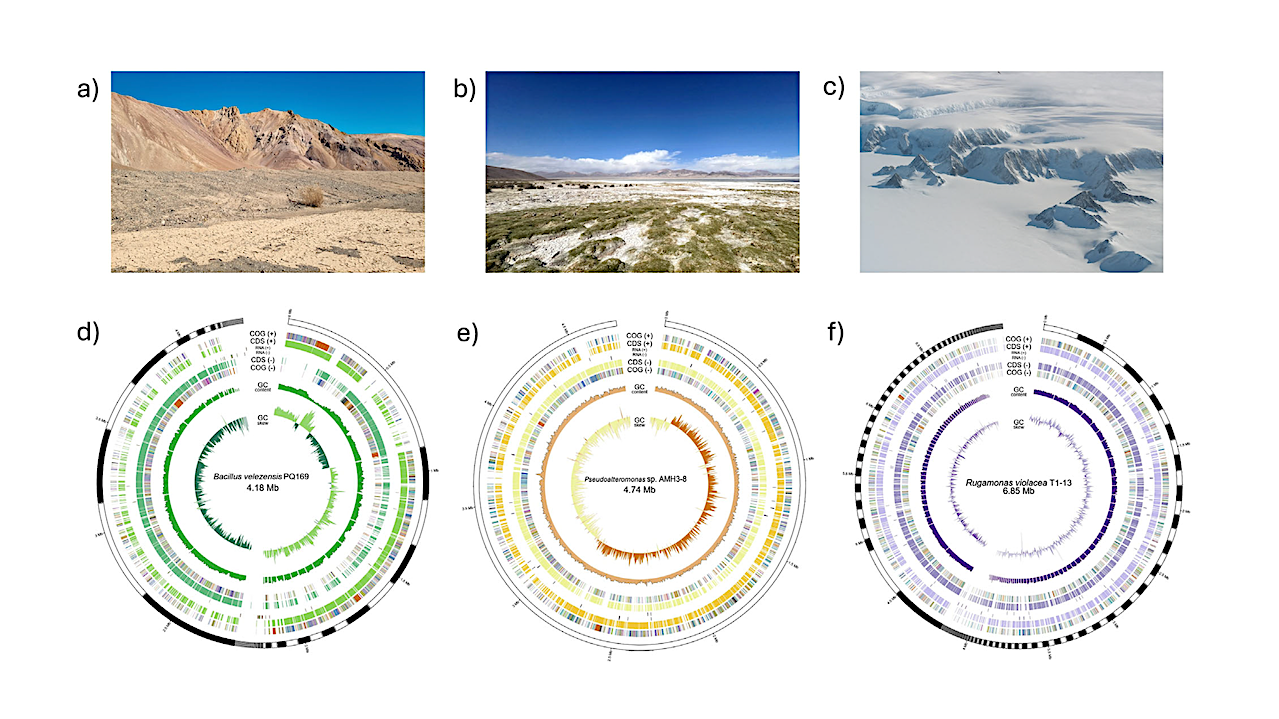
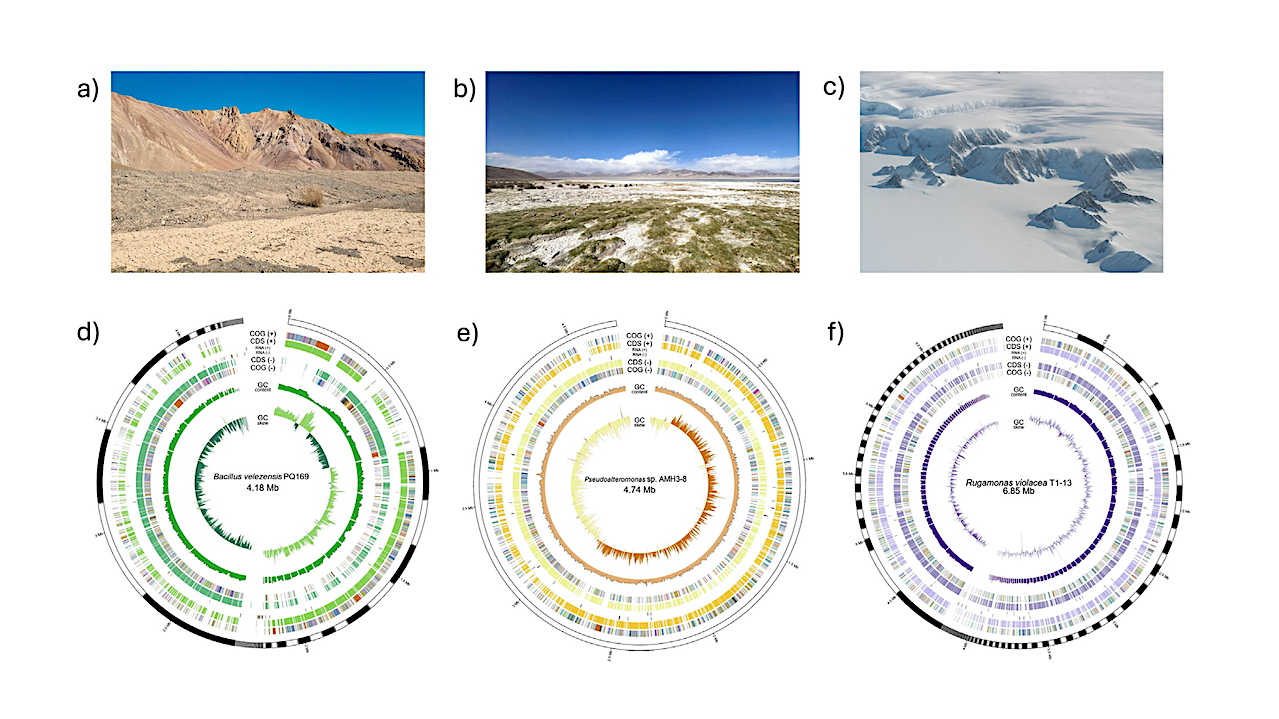
Isolation sites, genomic assembly characteristics, and ANI-based taxonomic assignment of three extremophilic bacterial isolates. (a–c) Representative sampling sites of bacterial isolates: (a) Atacama Desert, (b) Salar de Huasco, and (c) Antarctic soil. (d–f) Circular genome representations illustrating variability in genome size, contiguity, and genetic content: (d) Bacillus velezensis PQ169 (76 contigs, 4.18 Mb), (e) Pseudoalteromonas sp. AMH3-8 (single contig, 4.74 Mb), and (f) Rugamonas violacea T1-13 (171 contigs, 6.85 Mb). Rings (outer to inner) represent predicted CDSs, RNA genes (rRNA, tRNA, and tmRNA), GC content, and GC skew. — Int J Mol Sci via PubMed
Extremophilic bacteria from extreme environments, such as the Atacama Desert, Salar de Huasco, and Antarctica, exhibit adaptations to intense UV radiation. In this study, we investigated the genomic and structural mechanisms underlying UV resistance in three bacterial isolates identified as Bacillus velezensis PQ169, Pseudoalteromonas sp. AMH3-8, and Rugamonas violacea T1-13.
Through integrative genomic analyses, we identified key genes involved in DNA-repair systems, pigment production, and spore formation. Phylogenetic analyses of aminoacidic sequences of the nucleotide excision repair (NER) system revealed conserved evolutionary patterns, indicating their essential role across diverse bacterial taxa.
Structural modeling of photolyases from Pseudoalteromonas sp. AMH3-8 and R. violacea T1-13 provided further insights into protein function and interactions critical for DNA repair and UV resistance. Additionally, the presence of a complete violacein operon in R. violacea T1-13 underscores pigment biosynthesis as a crucial protective mechanism.
In B. velezensis PQ169, we identified the complete set of genes responsible for sporulation, suggesting that sporulation may represent a key protective strategy employed by this bacterium in response to environmental stress.
Our comprehensive approach underscores the complexity and diversity of microbial adaptations to UV stress, offering potential biotechnological applications and advancing our understanding of microbial resilience in extreme conditions.

Phylogenetic tree based on the amino acid sequences of UvrA, UvrB, and UvrC proteins (NER system). Multiple sequence alignment was performed using MUSCLE on the Phylogeny.fr platform, followed by tree construction with PhyML and visualization using TreeDyn. The resulting tree was further arranged and refined using the Interactive Tree of Life (iTOL v7) tool. The maximum likelihood method was employed with 1000 bootstrap replicates, where the node sizes reflect the level of bootstrap support. Each label includes the gene name, the bacterial species, and the corresponding NCBI accession number. Three environmental strains are highlighted: Bacillus velezensis PQ-169 (green), Pseudoalteromonas sp. AMH3-8 (yellow), and Rugamonas violacea T1-13 (purple). These strains, along with other extremophilic, UV-resistant, and pathogenic bacteria, were included to compare UvrA (red clade), UvrB (blue clade), and UvrC (green clade) variations under extreme environmental conditions. — Int J Mol Sci via PubMed
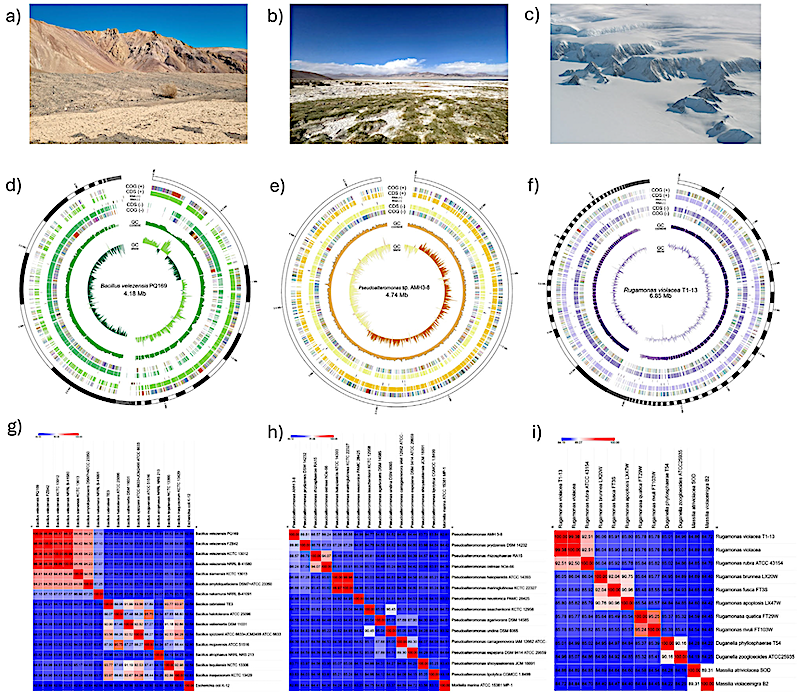
Isolation sites, genomic assembly characteristics, and ANI-based taxonomic assignment of three extremophilic bacterial isolates. (a–c) Representative sampling sites of bacterial isolates: (a) Atacama Desert, (b) Salar de Huasco, and (c) Antarctic soil. (d–f) Circular genome representations illustrating variability in genome size, contiguity, and genetic content: (d) Bacillus velezensis PQ169 (76 contigs, 4.18 Mb), (e) Pseudoalteromonas sp. AMH3-8 (single contig, 4.74 Mb), and (f) Rugamonas violacea T1-13 (171 contigs, 6.85 Mb). Rings (outer to inner) represent predicted CDSs, RNA genes (rRNA, tRNA, and tmRNA), GC content, and GC skew. (g–i) Heatmaps depicting average nucleotide identity based on MUMmer (ANIm) analysis between each isolate and closely related reference strains, supporting definitive taxonomic assignment: (g) Bacillus velezensis PQ169, (h) Pseudoalteromonas sp. AMH3-8, and (i) Rugamonas violacea T1-13. Color gradients indicate similarity percentages, with red color representing higher ANI values, and blue lower values. — Int J Mol Sci via PubMed
astrobiology, genomics, extremophiles,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly