Now Reading: Molecular Diversity As A Biosignature
-
01
Molecular Diversity As A Biosignature
Molecular Diversity As A Biosignature
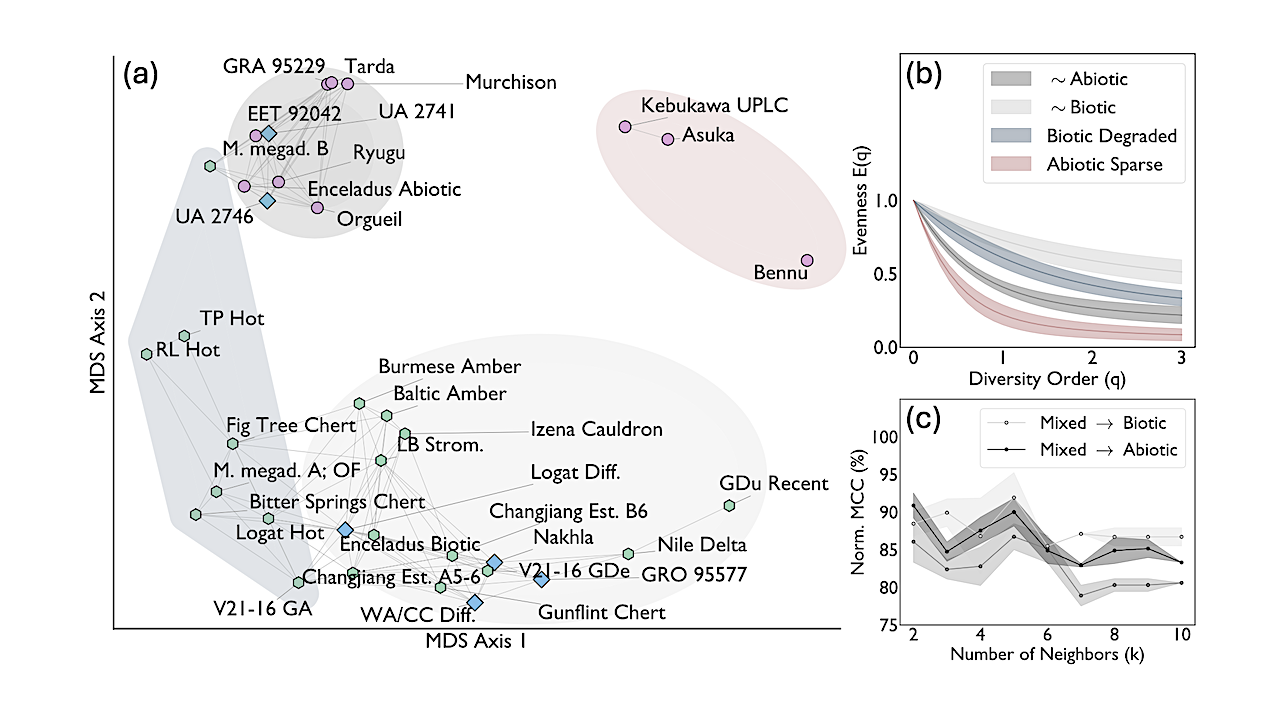
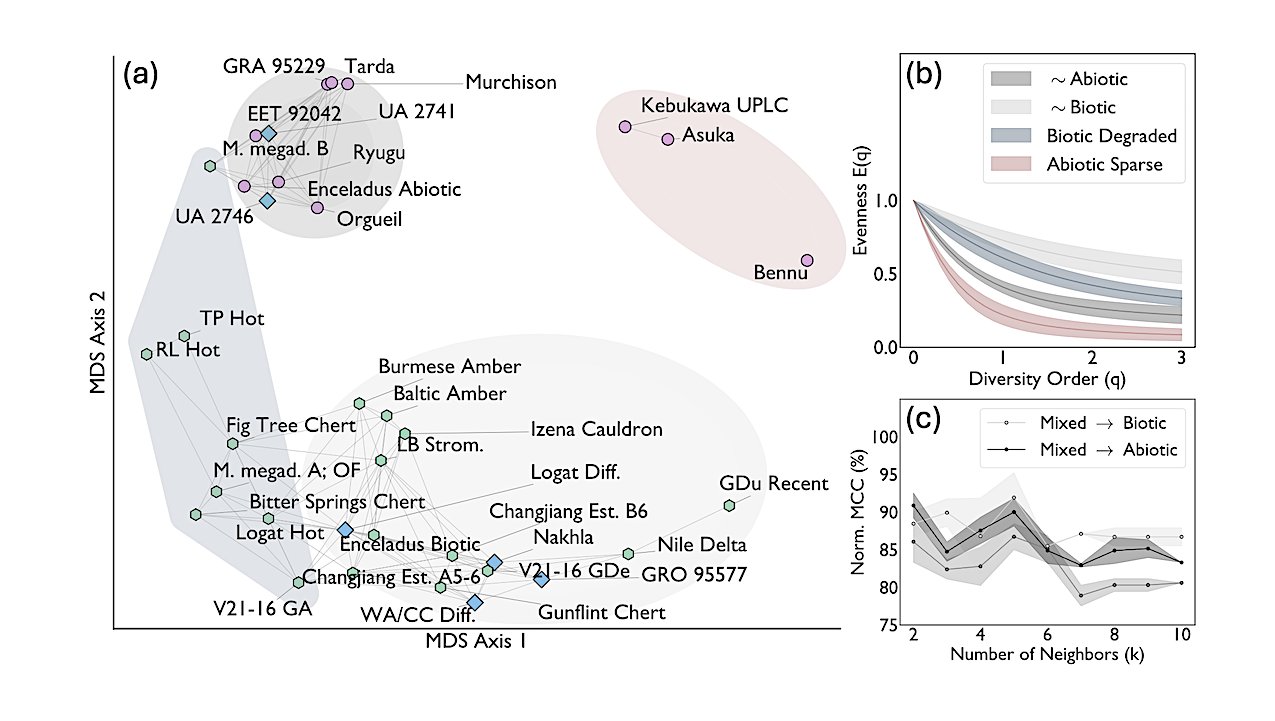
Dissimilarity analysis of evenness curves of amino acid assemblages. (a): Multidimensional Scaling (MDS) projection of dissimilarities between evenness curves, E(q). Points represent samples; distances between samples grow with dissimilarity. Edges connect samples to the 25th percentile of their nearest neighbors. Markers indicate inferred origin: biotic (green hexagons), abiotic (pink circles), and mixed (blue diamonds). (b): Evenness curve distributions of four distinct sample groups. Solid lines represent mean values, and filled areas represent the one standard deviation interval. (c): Predictive power of a sample’s origin through k-Nearest-Neighbors (kNN) classification, applied to pairwise distances between samples projected onto two MDS axes. Accuracy is quantified using a normalized Matthews Correlation Coefficient (MCC), where 50% corresponds to random assignment and 100% indicates perfect classification. Uncertainty was estimated using multiple initializations of the MDS projection. — astro-ph.EP
The search for life in the Solar System hinges on data from planetary missions. Biosignatures based on molecular identity, isotopic composition, or chiral excess require measurements that current and planned missions cannot provide.
We introduce a new class of biosignatures, defined by the statistical organization of molecular assemblages and quantified using ecodiversity metrics. Using this framework, we analyze amino acid diversity across a dataset spanning terrestrial and extraterrestrial contexts.
We find that biotic samples are consistently more diverse, and therefore distinct, from their sparser abiotic counterparts. This distinction holds for fatty acids as well, indicating that the diversity signal reflects a fundamental biosynthetic signature. It also proves persistent under space-like degradation.
Relying only on relative abundances, this biogenicity assessment strategy is applicable to any molecular composition data from archived, current, and planned planetary missions. By capturing a fundamental statistical property of life’s chemical organization, it may also transcend biosignatures that are contingent on Earth’s evolutionary history.
Gideon Yoffe, Fabian Klenner, Barak Sober, Yohai Kaspi, Itay Halevy
Subjects: Earth and Planetary Astrophysics (astro-ph.EP); Applications (stat.AP)
Cite as: arXiv:2511.00525 [astro-ph.EP] (or arXiv:2511.00525v1 [astro-ph.EP] for this version)
https://doi.org/10.48550/arXiv.2511.00525
Focus to learn more
Submission history
From: Gideon Yoffe
[v1] Sat, 1 Nov 2025 12:00:15 UTC (2,381 KB)
https://arxiv.org/abs/2511.00525
Astrobiology,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly