Now Reading: Organic Geochemical Evidence For Life In Archean Rocks Identified By Pyrolysis–GC–MS and Supervised Machine Learning
-
01
Organic Geochemical Evidence For Life In Archean Rocks Identified By Pyrolysis–GC–MS and Supervised Machine Learning
Organic Geochemical Evidence For Life In Archean Rocks Identified By Pyrolysis–GC–MS and Supervised Machine Learning
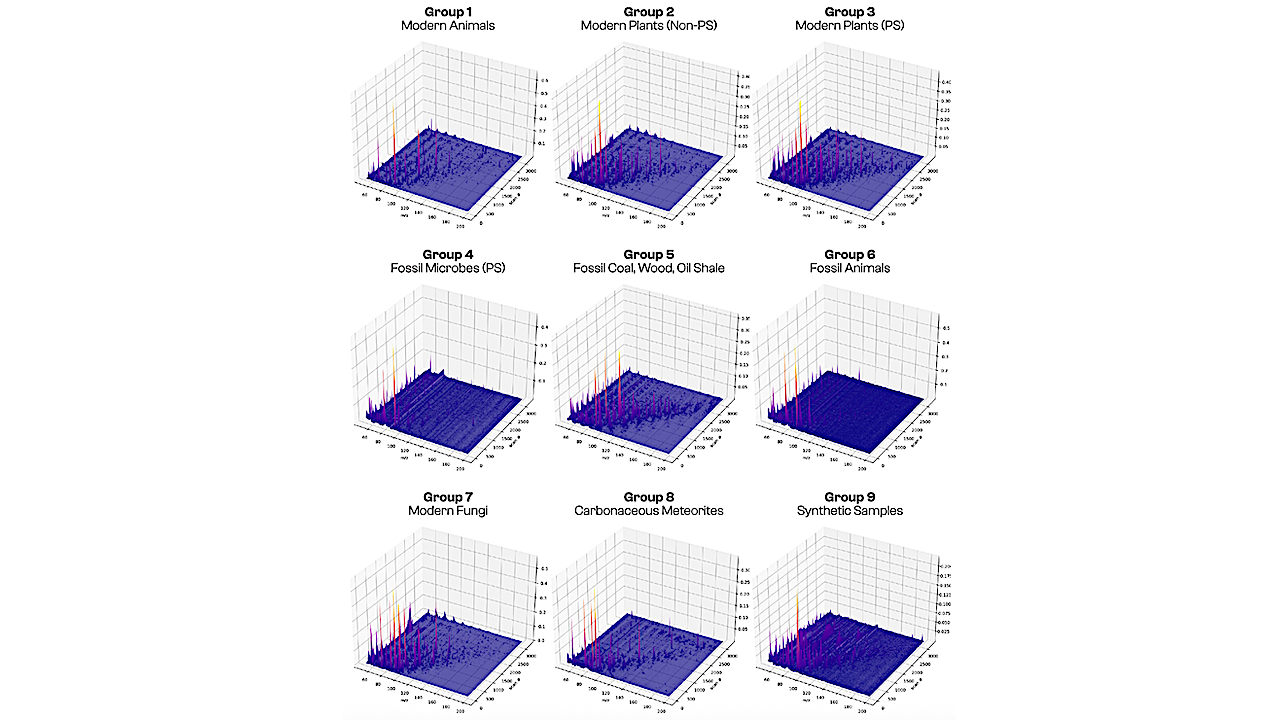
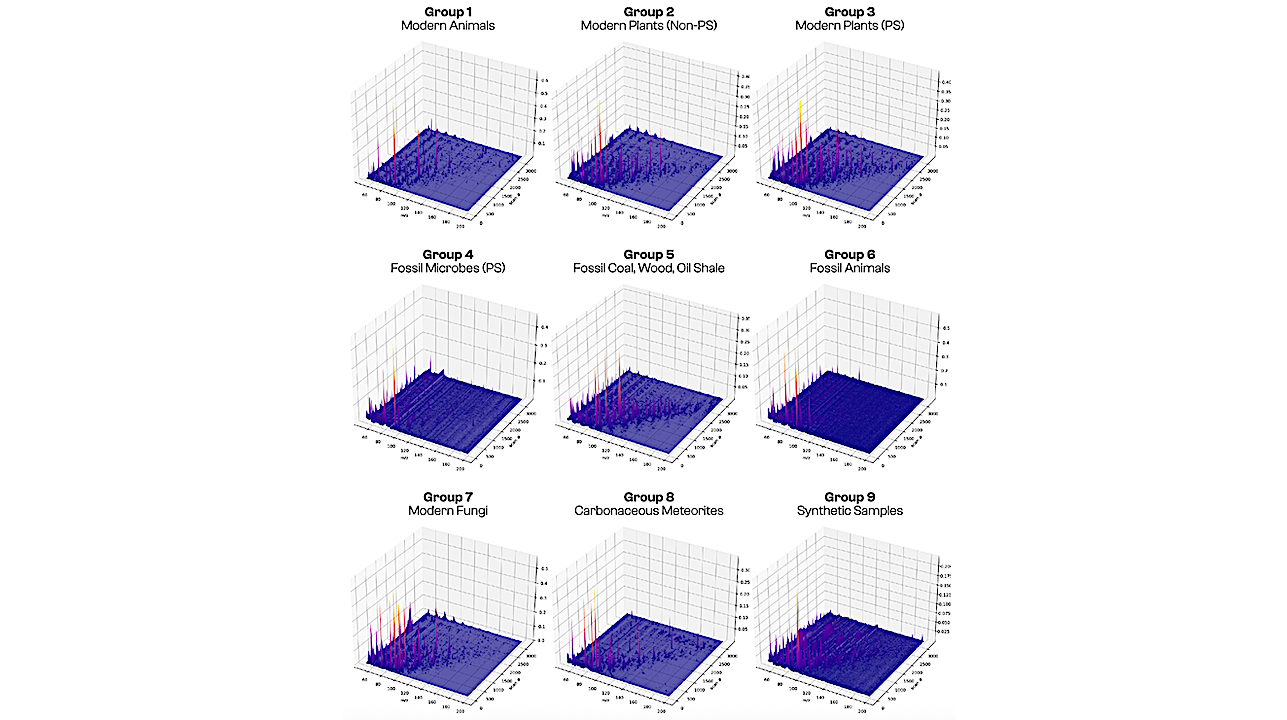
Aggregated three-dimensional py–GC–MS data for samples in each of our nine categories: modern animals; modern plants (non-photosynthetic); modern plants (photosynthetic); fossil microbes (photosynthetic); fossil coal, wood, oil shale; fossil animals; modern fungi; carbonaceous meteorites; and synthetic samples. These graphs display peak intensities (vertical scale, normalized to the highest peak intensity in each category) for 3,240 elution time bins or “scans” (right-hand scale) and their mass spectra over 150 m/z bins (left-hand scale). — PNAS
Significance
Teasing out biochemical information from ancient organic-rich sediments, notably the timing of the emergence of photosynthesis relative to the inferred oxygenation of Earth’s atmosphere, remains a challenging opportunity. To tackle this problem, we analyzed 406 diverse ancient and modern samples and used supervised machine learning to discriminate samples of biogenic vs. abiogenic origin, as well as photosynthetic vs. nonphotosynthetic physiology.
Comparing organic-rich samples of uncertain affinity to our training data, ca. 3.33-billion-year-old sedimentary rocks group among microbial samples, and rocks as old as 2.52 billion years ally with more recent photosynthetic life. The application of supervised machine learning thus approximately doubles the interval within which fossil organic matter can be shown to retain molecular information of evolutionary relationships and physiology.
Abstract
Throughout Earth’s history, organic molecules from both abiogenic and biogenic sources have been buried in sedimentary rocks. Most of these organic molecules have been significantly altered by geologic processes through deep time.
Nonetheless, the nature and distribution of those ancient fragmentary organic remains have the potential to reveal diagnostic biomolecular information after billions of years of burial. Here, we analyzed 406 fossil, modern biological, meteoritic, and synthetic samples using pyrolysis gas chromatography and mass spectrometry.
We explored these analytical data via supervised machine-learning methods to discriminate samples of biogenic vs. abiogenic origin, plant vs. animal phylogenetic affinity, and photosynthetic vs. nonphotosynthetic physiology.
Dividing 272 samples with known phylogenetic affinity and physiology into 9 categories, each further divided into 75% training and 25% testing sets, our random forest models accurately predict pairwise assignments of modern vs. fossil or meteoritic organics (100% correct assignments), fossil plant tissues vs. meteoritic organics (97%), modern vs. fossil plant tissues (98%), and modern plants vs. animal tissues (95%). Pairwise comparisons between fossil biogenic samples vs. abiogenic samples resulted in 93% correct classifications, while analysis of modern and ancient photosynthetic vs. nonphotosynthetic samples also resulted in 93% correct assignments.
Our analyses demonstrate that molecular biosignatures can survive in ancient fossils and allow for the identification of organismal origins and traits. Consistent with previous morphological and isotopic inferences, we present evidence for biogenic molecular assemblages in Paleoarchean rocks (3.33 Ga) and for photoautotrophy in Neoarchean rocks (2.52 Ga).
Organic geochemical evidence for life in Archean rocks identified by pyrolysis–GC–MS and supervised machine learning, PNAS (open access)
Astrobiology, Astrogeology,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly