Now Reading: The Eukaryotic Cell Emerged As An Evolutionary Algorithmic Phase Transition
-
01
The Eukaryotic Cell Emerged As An Evolutionary Algorithmic Phase Transition
The Eukaryotic Cell Emerged As An Evolutionary Algorithmic Phase Transition
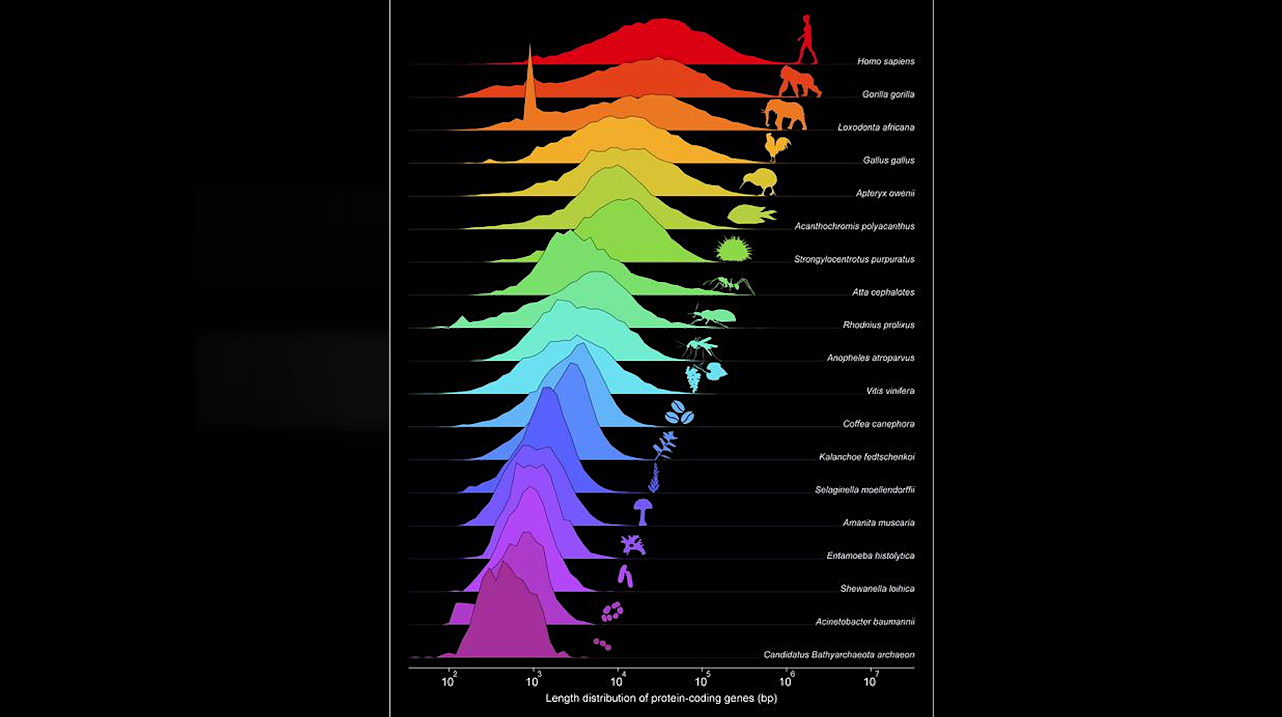

Evolution of protein-coding gene length distributions across evolutionary time Credit ill./©: Fernando J. Ballesteros
An international collaboration between four senior scientists from Mainz, Valencia, Madrid, and Zurich has published groundbreaking research in the journal PNAS, shedding light on the most significant increase in complexity in the history of life’s evolution on Earth: the origin of the eukaryotic cell.
While the endosymbiotic theory is widely accepted, the billions of years that have passed since the fusion of an Archaea and a Bacteria have resulted in a lack of evolutionary intermediates in the phylogenetic tree until the emergence of the eukaryotic cell. It is a gap in our knowledge, referred to as the black hole at the heart of biology.
“The new study is a blend of theoretical and observational approaches that quantitatively understands how the genetic architecture of life was transformed to allow such an increase in complexity”, stated Dr. Enrique M. Muro, representative of Johannes Gutenberg University Mainz (JGU) in this project.
Proteins and protein coding genes increase in length
The article in PNAS demonstrates that the distributions of protein lengths and their corresponding genes follow log-normal distributions across the whole tree of life. To do this, 9,913 different proteomes and 33,627 genomes were analyzed. Log-normal distributions typically arise as a result of multiplicative processes. Following Ockham’s razor principle, the researchers modeled the evolution of gene length distributions as multiplicative stochastic processes.
In fact, they modeled the action of all genetic operators combined in relation to sequence length. Starting from LUCA, i.e., the hypothesized last universal common ancestor from which the three domains of life – the Bacteria, the Archaea, and the Eukarya – originated, the researchers found both theoretically and observationally that the average gene lengths have evolved exponentially over evolutionary time across different species.
Furthermore, they discovered a scaling-invariant mechanism of gene growth across the entire tree of life, where the variance directly depends on the mean protein length. By representing all the species captured in the 33,627 genomes, the team was able to observationally verify the predictions and, moreover, show that the average gene length is a very good surrogate for organismal complexity.
In a pure exercise of quantitative biology, Dr. Bartolo Luque from the Polytechnic University of Madrid added: “From knowing the average length of protein-coding genes in a species, we can calculate the whole distribution of gene length within that species.”
When representing the evolution of the average protein lengths versus their corresponding gene lengths across different species, it is observed that they evolve simultaneously in prokaryotes, because there are almost no non-coding sequences in their genes. However, once the average gene length reaches 1,500 nucleotides, the proteins decouple from the multiplicative process of gene growth, and the average protein length stabilizes after the onset of the eukaryotic cell at about 500 amino acids in a clear threshold, marking the appearance of the eukaryotic cell. From that point onward, and unlike what happens with proteins, the average gene length continues to increase as it did in prokaryotes, due to the presence of non-coding sequences.

Scale-invariant relationship between mean gene length and variance. Each dot in this log–log plot represents the genome of a single species ( 33,627), with H. sapiens highlighted by the circle. Gene length is measured in number of base pairs. Different colors identify major phyletic groups. As a guide to the eye, we have added the labels fishes, Aves, and Primates, marking the region where these groups cluster. The line represents the best fit to a power law (Eq. 4; , ). Inset: equivalent representation for protein length (measured in number of amino acids) for 9,913 species. The line represents the best fit to an equivalent power law (SI Appendix; , ). Major phyletic groups appear sequentially across the scaling law for genes, but not so clearly in the equivalent for proteins. – -PNAS
Algorithmic phase transition
A critical phenomena analysis then concluded that a phase transition, well studied in the physics of magnetic materials, occurred at a critical gene length of 1,500 nucleotides. This marked eukaryogenesis and divides the evolution of life into two distinct phases: a coding phase (Prokarya) and a non-coding phase (Eukarya).
Additionally, characteristic phenomena of these transitions are observed, such as critical slowing down, where the system’s dynamics become trapped in many metastable states around the critical point. “This is corroborated in early protists and fungi”, said Dr. Fernando Ballesteros from the University of Valencia.
Moreover, “the phase transition was algorithmic,” added Professor Jordi Bascompte from the University of Zurich. In the coding phase, in a scenario close to LUCA, with short proteins, increasing the length of proteins and their corresponding genes was computationally simple.
However, as the protein lengths grew, the search for longer proteins became unfeasible. This tension caused by genes that grew at the same rate as before while proteins could not was resolved continuously but abruptly with the incorporation of non-coding sequences into the genes.
With this innovation, the algorithm for searching for new proteins rapidly reduced its computational complexity, becoming non-linear through the spliceosome and the nucleus, which separated transcription and splicing from translation. This happened at the critical point of phase transition, which this study dates to 2.6 billion years ago.
The study recently published in PNAS not only answers essential questions, but is interdisciplinary, combining computational biology, evolutionary biology, and physics. “It has the potential to interest a wide audience across many disciplines and serve as a foundation for other groups to explore different research avenues, such as energy or information theory”, emphasized Dr. Enrique Muro of the Institute of Organismic and Molecular Evolution at Mainz University.
The eukaryotic cell, the most significant increase in complexity in the history of life’s evolution on Earth, emerged as a phase transition and unlocked the path toward other major transitions – such as multicellularity, sexuality, and sociability – that shaped life on our planet as we know it today.
The emergence of eukaryotes as an evolutionary algorithmic phase transition, Proceedings of the National Academy of Sciences (open access)
Astrobiology,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly