Now Reading: What Lives? A Meta-analysis Of Diverse Opinions On The Definition Of Life
-
01
What Lives? A Meta-analysis Of Diverse Opinions On The Definition Of Life
What Lives? A Meta-analysis Of Diverse Opinions On The Definition Of Life
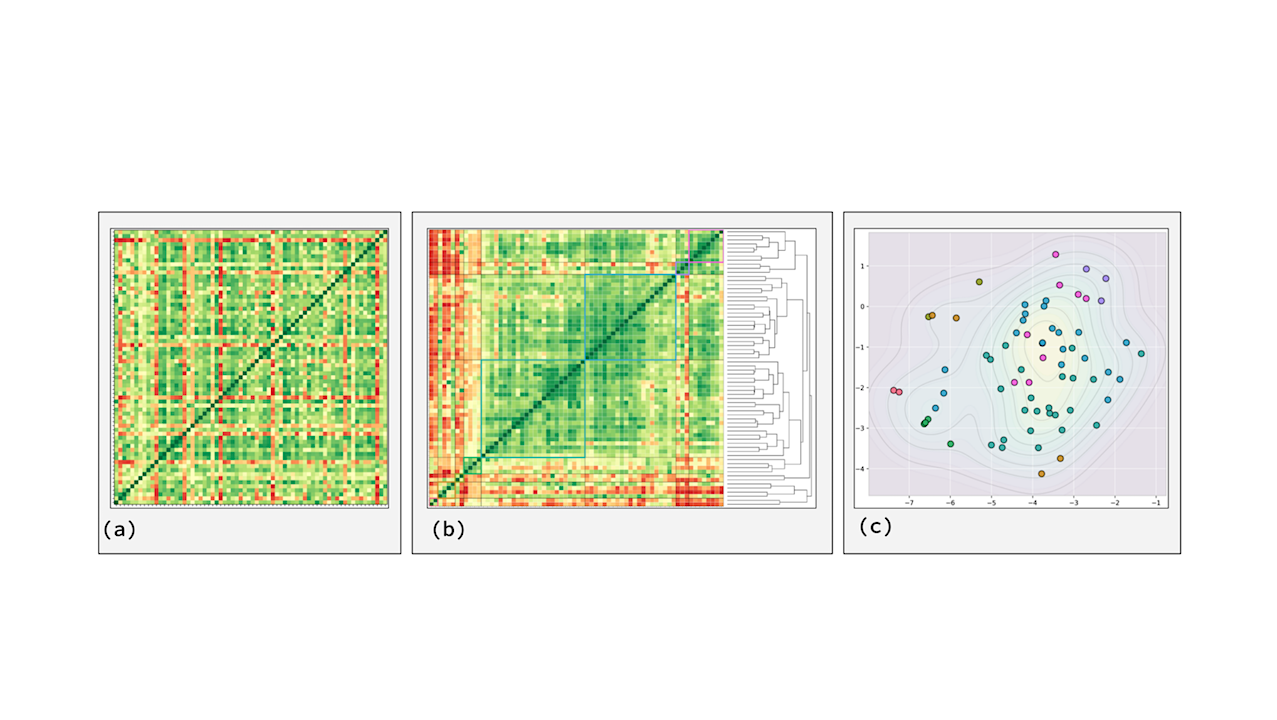

Data transformation process from pairwise correlations to clustered spatial projection. Panel (a) shows the initial pairwise correlation matrix for all 68 definitions, with green indicating high agreement (correlation approaching 1.0) and red indicating disagreement (correlation approaching -1.0). Panel (b) illustrates the sorted correlation matrix, produced after applying hierarchical clustering, with the resulting dendrogram displayed on the right showing the nested relationships between definitions. Panel (c) presents the final t-SNE projection that transforms these high-dimensional correlation patterns into an interpretable 2D semantic landscape, with colors indicating cluster membership and contour lines revealing density variations across the definitional space. — q-bio.OT
The question of “what is life?” has challenged scientists and philosophers for centuries, producing an array of definitions that reflect both the mystery of its emergence and the diversity of disciplinary perspectives brought to bear on the question.
Despite significant progress in our understanding of biological systems, psychology, computation, and information theory, no single definition for life has yet achieved universal acceptance. This challenge becomes increasingly urgent as advances in synthetic biology, artificial intelligence, and astrobiology challenge our traditional conceptions of what it means to be alive.
We undertook a methodological approach that leverages large language models (LLMs) to analyze a set of definitions of life provided by a curated set of cross-disciplinary experts. We used a novel pairwise correlation analysis to map the definitions into distinct feature vectors, followed by agglomerative clustering, intra-cluster semantic analysis, and t-SNE projection to reveal underlying conceptual archetypes.
This methodology revealed a continuous landscape of the themes relating to the definition of life, suggesting that what has historically been approached as a binary taxonomic problem should be instead conceived as differentiated perspectives within a unified conceptual latent space.
We offer a new methodological bridge between reductionist and holistic approaches to fundamental questions in science and philosophy, demonstrating how computational semantic analysis can reveal conceptual patterns across disciplinary boundaries, and opening similar pathways for addressing other contested definitional territories across the sciences.
Reed Bender, Karina Kofman, Blaise Agüera y Arcas, Michael Levin
Comments: 54 pages, 4 figures, 2 tables, 11 supplemental figures, 3 supplemental tables
Subjects: Other Quantitative Biology (q-bio.OT); Artificial Intelligence (cs.AI); Computers and Society (cs.CY); Biomolecules (q-bio.BM); Cell Behavior (q-bio.CB); Subcellular Processes (q-bio.SC); Applications (stat.AP)
Cite as: arXiv:2505.15849 [q-bio.OT] (or arXiv:2505.15849v1 [q-bio.OT] for this version)
https://doi.org/10.48550/arXiv.2505.15849
Focus to learn more
Submission history
From: Michael Levin
[v1] Mon, 19 May 2025 20:17:37 UTC (7,499 KB)
https://arxiv.org/abs/2505.15849
Astrobiology,
Stay Informed With the Latest & Most Important News
Previous Post
Next Post
-
 01Two Black Holes Observed Circling Each Other for the First Time
01Two Black Holes Observed Circling Each Other for the First Time -
 02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life
02From Polymerization-Enabled Folding and Assembly to Chemical Evolution: Key Processes for Emergence of Functional Polymers in the Origin of Life -
 03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series)
03Astronomy 101: From the Sun and Moon to Wormholes and Warp Drive, Key Theories, Discoveries, and Facts about the Universe (The Adams 101 Series) -
 04True Anomaly hires former York Space executive as chief operating officer
04True Anomaly hires former York Space executive as chief operating officer -
 05Φsat-2 begins science phase for AI Earth images
05Φsat-2 begins science phase for AI Earth images -
 06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters
06Hurricane forecasters are losing 3 key satellites ahead of peak storm season − a meteorologist explains why it matters -
 07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly
07Binary star systems are complex astronomical objects − a new AI approach could pin down their properties quickly